We are investigators at the Center for Computation and Technology (CCT), LSU. Our research interest is to develope high-performance and distributed computing strategies for large scale scientfic applications. In particular, biological applications such as Next-generation sequencing analytics, molecular simulations, and advanced sampling are the major topics aimed by the developed strategies. HPC has been a key role for supporting large scale applications, and distributed computing strategies have been increasingly becoming critical, in particular, data intensive applications and BigData challenges. Currently, we focus on specific reesearch topics described below. You can contact us with e-mail. jhkim@cct.lsu.edu, nykim@cct.lsu.edu
Science gateways are effective means for collaborative research activities with the capability to utilize large scale High-performance computing (HPC) resources and databases located in many different places. Gateways lower significantly barriers associated with all kinds of complexities associated with accessing and such resources and data sets, and eventually facilitate scientific discovery of an individual researcher. While most of successful gateways are concerned about large scale compute intensive scientific applications, gateway is also an attractive method for BigData projects. We have been developing a gateway architecture along with collaborators and the schematic is shown here. Our approach provides an effective way to develop a new science gateway which is capable of the seamless integration of distributed resources and databases, while the development cycle is reletively shorter due to our design strategy to build interoperable, distributed, adative, and user-friendly research cyberinfrastructure.
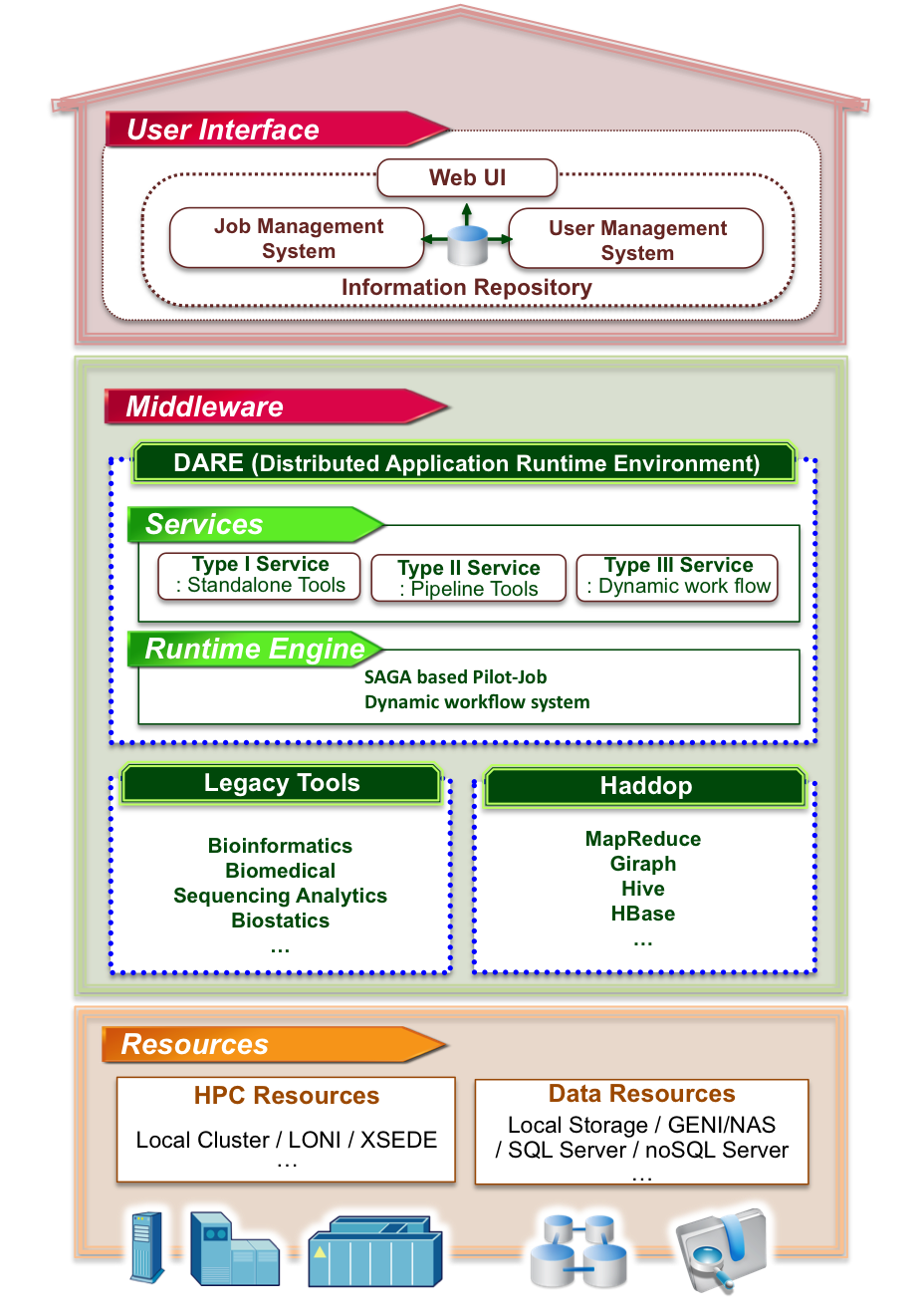
Sharath Maddineni, Joohyun Kim, Yaakoub El-Khamra, and Shantenu Jha, “Dynamic Application Runtime Environment (DARE): A Standards-based Framework For Building Science Gateways”, J. Grid Computing, (2012), 10(4):647
Our research interest is to develop novel strategies for large scale bigdata genomics projects. Some major focus areas are stated below.
Large scale simulations of biomolecules, in particular, RNAs have been pursued. Our strategy is to utilize High-performance Computing (HPC) machines as well as to develop efficient sampling methods such as Replica Exchange Statistical Temperature Molecular Dynamics. Some of our recent contributions, along with collaborators, are as follow
We are currently engaged in genelab NGS sequencing core, BIOMMED, School of Vet. Medicine, LSU as a bioinformatics support team. For more information, visit this link. The overall infrastructure behind the sequencing core and bioinformatics support can be illustrated as follows.
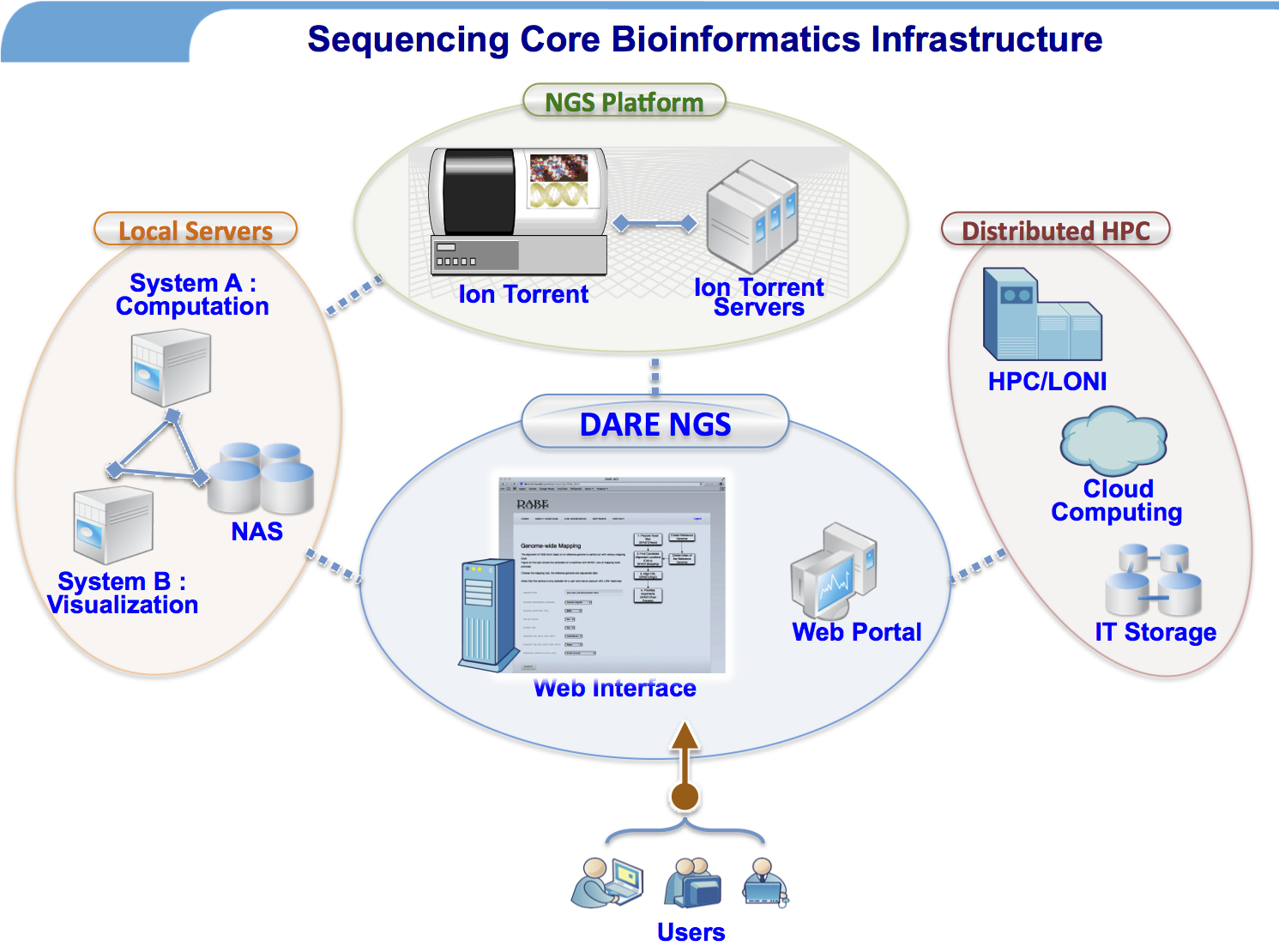
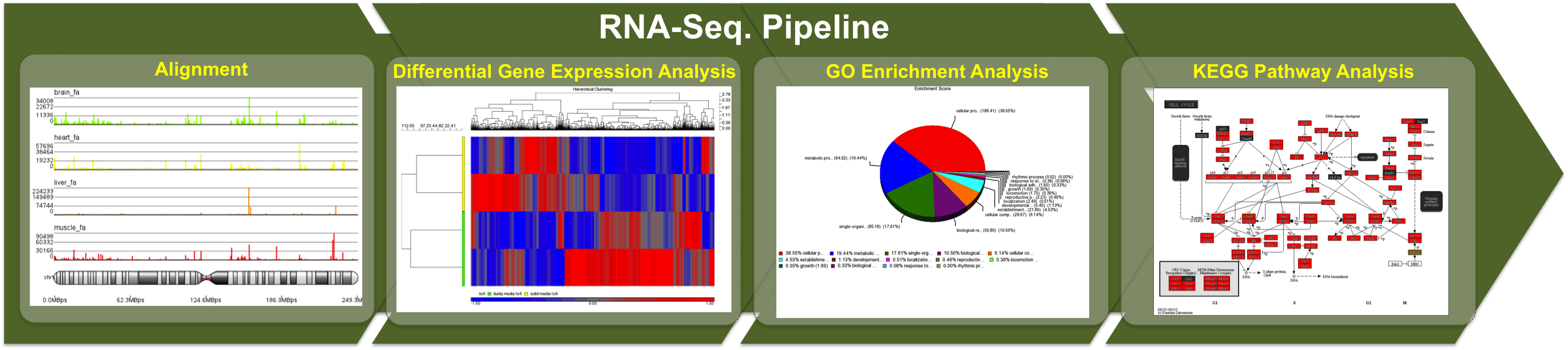
Joohyun Kim, Sharath Maddineni, and Shantenu Jha, “Advancing Next-Generation Sequencing Data Analytics with Scalable Distributed Infrastructure”, (published online with Concurrency and Computation: Practice and Experience).
Felix Francis, Joohyun Kim, Thiru Ramaraj, Andrew Farmer, Milton C. Rush, and Jong Hyun Ham, “Comparative genomic analysis of two Burkholderia glumae strains from different geographic origins reveals a high degree of plasticity in genome structure associated with genomic islands” , Mol. Genet. Genomics, (2013) 288:195–203.